|
Imperial College Computational Bioinformatics Laboratory (CBL)
|
The CBL is physically situated in the Department of Computing in what
is presently Room 407 Huxley (an area containing 3 offices and a large open
area for post-docs and PhD students).
Permanent staff include Prof Stephen Muggleton (Joint Research Chair
in Computational Bioinformatics),
Dr Simon Colton (Lecturer)
and Janice Lonsdale (secretary).
The remit of the Centre is the study of the theory, implementation
and application of computational techniques to problems in Biology
and Medicine. The Centre is developing strong and growing links with
other related groups and centres across Imperial College.
Research
This page contains:-
-
an overview of scientific knowledge discovery tasks carried
out using Inductive Logic Programming (ILP);
-
a bibliography of the corresponding publications;
-
links to other pages on current and proposed projects.
The results reviewed
have been published in some of the top general science journals, and
as such are among the strongest examples of semi-automated
scientific discovery in the Artificial Intelligence literature.
Scientific Knowledge Discovery using ILP
The pharmaceutical industry is increasingly overwhelmed by
large-volume-data. This is generated both internally as a side-effect
of screening tests and combinatorial chemistry, as well as externally
from sources such as the human genome project. On the other hand the
industry is predominantly knowledge-driven. For instance, knowledge
is required within computational chemistry for pharmacophore
identification, as well as for determining biological function using
sequence analysis.
From a computer science point of view, the knowledge requirements
within the industry give higher emphasis to ``knowing that''
(declarative or descriptive knowledge) rather than ``knowing how''
(procedural or prescriptive knowledge). Mathematical logic has always
been the preferred representation for declarative knowledge and thus
knowledge discovery techniques are required which generate logical
formulae from data. Inductive Logic Programming (ILP) [8,1] provides such an
approach.
ILP algorithms take examples E of a concept (such as a
protein family) together with background knowledge B (such as
a definition of molecular dynamics) and construct a hypothesis
H which explains E in terms of B. For
example, in protein fold domains, E might consist of
descriptions of molecules separated into positive and negative
examples of a particular fold (overall protein shape). This is
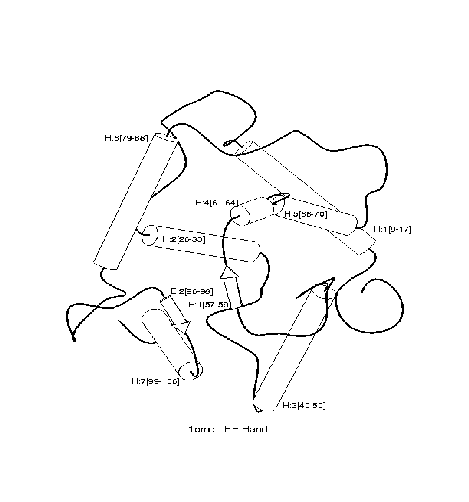
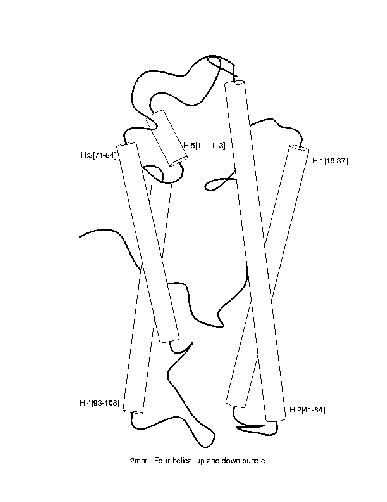
exemplified in the diagram below for the fold
``4-helical-up-and-down-bundle''.
Positive and negative examples of a ``4-helical-up-and-down-bundle''.
|
Positive
|
Negative
|
|
|
A possible hypothesis H describing this class of proteins is:
fold('Four-helical up-and-down bundle',P) :-
helix(P,H1),
length(H1,hi),
position(P,H1,Pos),
interval(1 =< Pos =< 3),
adjacent(P,H1,H2),
helix(P,H2).
The hypothesis is a definite clause consisting of a head
(fold(..,..)) and a body (the conjunction length(..),
.. helix(..)). In this case ``fold'' is the predicate involved in the
examples and hypothesis, while ``length'', ``position'', etc. are
defined by the background knowledge. A logic program is simply a set
of such definite clauses. Each of E, B and
H are logic programs.
In the context of knowledge discovery a distinct advantage of ILP
over black box techniques, such as neural networks, is that a
hypotheses such as that shown above can, in a straightforward manner,
be automatically translated into the following piece of English text.
The protein P has fold class ``Four-helical up-and-down bundle''
if it contains a long helix H1 at a secondary structure position
between 1 and 3, and H1 is followed by a second helix H2.
Such explicit hypotheses can be used within the familiar human
scientific discovery cycle of debate, criticism and refutation.
Discovery of biological function
Biological functions are regulated by the docking of small molecules
(ligands) with sites on large molecules (proteins). Drugs, such as
beta-blockers, mimic natural small molecules, such as adrenaline.
Effectiveness of drugs depends on the correct shape and charge
distribution of ligands. Thus beta-blockers block the binding of
adrenaline, and so stop over-stimulation of heart muscle in patients
prone to heart attacks.
Results on scientific discovery applications of ILP are separated
below between those related to small molecules (such as ligands)
and those related to proteins.
Small molecules
Structure-activity prediction
The majority of pharmaceutical R&D is based on finding slightly
improved variants of patented active drugs. This involves
laboratories of chemists synthesising and testing hundreds of
compounds almost at random. The average cost of developing a single
new drug is around $300 million. In [3] it was shown that ILP system
Golem [6] was capable of
constructing rules which accurately predict the activity of untried
drugs. Rules were constructed from examples of drugs with known
medicinal activity. The accuracy of the rules was found to be slightly
higher than traditional statistical methods. More importantly the
easily understandable rules provided insights which were directly
comparable to the relevant literature concerning the binding site of
dihydrofolate reductase.
Mutagenesis
In [4],[10] ILP system Progol [5] was used to predict the
mutagenicity of chemical compounds taken from a previous study in
which linear regression had been applied. Progol's predictive accuracy
was equivalent to regression on the main set of 188 compounds and
significantly higher (85.7% as opposed to 66.7%) on 44 compounds which
had been discarded by the previous authors as unpredictable using
regression. Progol's single clause solution for the 44 compounds was
judged by the domain experts to be a new structural alert for
mutagenesis.
Pharmacophores
In a series of ``blind tests'' in collaboration with the
pharmaceutical company Pfizer UK, Progol was shown [2]
capable of re-discovering a 3D description of the binding sites (or
pharmacophores) of ACE inhibitors (a hypertension drug) and an
HIV-protease inhibitor (an anti-AIDS drug).
Carcinogenicity
Last year Progol was entered into a world-wide carcinogenicity
prediction competition run by the National Toxicology Program (NTP) in
the USA. Progol was trained on around 300 available compounds, and
made use of its earlier rules relating to mutagenicity. In the first
round of the competition Progol produced the highest predictive
accuracy of any automatic system entered [9].
Comparative accuracies on the first round of the Predictive Toxicology
Evaluation (PTE-1). Here P represents the binomial
probability that Progol and the corresponding toxicity prediction
method classify the same proportion of examples correctly. The
``Default'' method predicts all compounds to be carcinogenic. Methods
marked with a * have access to short-term in vivo rodent tests
that were unavailable to other methods. Ashby and RASH also involve
some subjective evaluation to decide on structural alerts.
| Method | Type | Accuracy | P |
| Ashby* | Chemist | 0.77 | 0.29 |
| Progol | ILP | 0.72 | 1.00
|
| RASH* | Biological potency analysis | 0.72 | 0.39 |
| TIPT* | Propositional ML | 0.67 | 0.11 |
| Bakale | Chemical reactivity analysis | 0.63 | 0.09 |
| Benigni | Expert-guided regression | 0.62 | 0.02 |
| DEREK | Expert system | 0.57 | 0.02 |
| TOPKAT | Statistical discrimination | 0.54 | 0.03 |
| CASE | Statistical correlation analysis | 0.54 | < 0.01 |
| COMPACT | Molecular modelling | 0.54 | 0.01 |
| Default | Majority class | 0.51 | 0.01 |
Proteins
Protein secondary structure prediction.
In [7] Golem was applied to
one of the hardest open problems in molecular biology. The problem is
as follows: given a sequence of amino acid residues, predict the
placement of the main three dimensional sub-structures of the protein.
The problem is of great interest to pharmaceutical companies involved
with drug design. For this reason, over the last 20 years many
attempts have been made to apply methods ranging from statistical
regression to decision tree and neural net learning to this
problem. Published accuracy results for the general prediction problem
have ranged between 50 and 60%, very close to majority-class
prediction rates. In our investigation we found the ability to make
use of background knowledge from molecular biology, together with the
ability to describe structural relations boosted the predictivity for
a restricted sub-problem to around 80% on an independently chosen test
set.
Discovery of fold descriptions
Protein shape is usually described at various levels of abstraction. At
the lower levels each family of proteins contains members with high
sequence similarity. At the most abstract level folds describe
proteins which have similar overall shape but are very different at
the sequence level. The lack of understanding of shape determination
has made protein fold prediction particularly hard. However, it is
intriguing that although there are around 300 known folds, around half
of all known proteins are members of the 20 most populated folds. In
[12] Progol was applied to
discover rules governing these 20 most populated protein folds.
Average in class cross-validated prediction was around 70% and many of
the rules were judged to be good characterisations of the fold classes
by Michael Sternberg, a world-class protein prediction expert at the
Imperial Cancer Research Fund in London.
Conclusion
In his statement of the importance of this line of research to the
Royal Society [11] Sternberg
emphasised the aspect of joint human-computer collaboration in
scientific discoveries. Science is an activity of human societies.
It is our belief that computer-based scientific discovery must support
strong integration into existing the social environment of human
scientific communities. The discovered knowledge must add to and build
on existing science. We believe that the ability to incorporate
background knowledge and re-use learned knowledge together with the
comprehensibility of the hypotheses, have marked out ILP as a
particularly effective approach for scientific knowledge
discovery.
Bibliography
-
- 1
-
I. Bratko and S. Muggleton.
Applications of inductive logic programming.
Communications of the ACM, 38(11):65-70, 1995.
- 2
-
P. Finn, S. Muggleton, D. Page, and A. Srinivasan.
Pharmacophore discovery using the inductive logic programming system
Progol.
Machine Learning, 30:241-271, 1998.
- 3
-
R. King, S. Muggleton, R. Lewis, and M. Sternberg.
Drug design by machine learning: The use of inductive logic
programming to model the structure-activity relationships of trimethoprim
analogues binding to dihydrofolate reductase.
Proceedings of the National Academy of Sciences,
89(23):11322-11326, 1992.
- 4
-
R. King, S. Muggleton, A. Srinivasan, and M. Sternberg.
Structure-activity relationships derived by machine learning: the use
of atoms and their bond connectives to predict mutagenicity by inductive
logic programming.
Proceedings of the National Academy of Sciences, 93:438-442,
1996.
- 5
-
S. Muggleton.
Inverse entailment and Progol.
New Generation Computing, 13:245-286, 1995.
- 6
-
S. Muggleton and C. Feng.
Efficient induction of logic programs.
In S. Muggleton, editor, Inductive Logic Programming, pages
281-298. Academic Press, London, 1992.
- 7
-
S. Muggleton, R. King, and M. Sternberg.
Protein secondary structure prediction using logic-based machine
learning.
Protein Engineering, 5(7):647-657, 1992.
- 8
-
S. Muggleton and L. De Raedt.
Inductive logic programming: Theory and methods.
Journal of Logic Programming, 19,20:629-679, 1994.
- 9
-
A. Srinivasan, , R.D. King S.H. Muggleton, and M. Sternberg.
Carcinogenesis predictions using ILP.
In N. Lavrac and S. Dzeroski, editors, Proceedings of
the Seventh International Workshop on Inductive Logic Programming, pages
273-287. Springer-Verlag, Berlin, 1997.
LNAI 1297.
- 10
-
A. Srinivasan, S. Muggleton, R. King, and M. Sternberg.
Theories for mutagenicity: a study of first-order and feature based
induction.
Artificial Intelligence, 85(1,2):277-299, 1996.
- 11
-
M. Sternberg, R. King, R. Lewis, and S. Muggleton.
Application of machine learning to structural molecular biology.
Philosophical Transactions of the Royal Society B,
344:365-371, 1994.
- 12
-
M. Turcotte, S.H. Muggleton, and M.J.E. Sternberg.
Protein fold recognition.
In C.D. Page, editor, Proc. of the 8th International Workshop
on Inductive Logic Programming (ILP-98), LNAI 1446, pages 53-64, Berlin,
1998. Springer-Verlag.