LEAP
In LEAP, manifold learning is used to boost multi-atlas segmentation by Learning Embeddings for Atlas Propagation. The method automatically learns a set of structural labels for a large and diverse image population by propagating manually defined label maps from a subset of the population to the entire image set. The automatically generated atlas database can be used to accurately label unseen images by selecting the most appropriate atlases from the entire pool. The resulting structural segmentation allows the accurate extraction of the volumes of different brain structures. 4D graph-cuts, an extension to LEAP, furthermore generates an accurate segmentation of longitudinal image sequences, allowing the reliable measurement of atrophy. The LEAP technology is licensed to IXICO Ltd. and currently tested for the application of measuring hippocampal volume in clinical trials of several major pharmaceutical companies.



Related Publications:
-
• R. Wolz, P. Aljabar, J.V. Hajnal, A. Hammers, D. Rueckert
LEAP: Learning Embeddings for Atlas Propagation
NeuroImage, 49(2):1316-1325, 2010
-
• R. Wolz, R.A. Heckemann, P. Aljabar, J.V. Hajnal, A. Hammers, J. Lotjonen and D. Rueckert
Measurement of hippocampal atrophy using 4D graph-cut segmentation: Application to ADNI
NeuroImage, 52(1):109-118, 2010
-
• L. Clerx, L. van der Pol, D. Rueckert, R. de Jong, R. v Schijndel et al.
Comparison of measurements of medial temporal lobe atrophy in the prediction of Alzheimer's Disease in subjects with MCI
Alzheimer's and Dementia, Volume 7, Issue 4, Supplement 1, July 2011, Page S133
-
• R. Wolz, D. Rueckert
Segmentation of the ADNI database into 83 anatomical regions using LEAP
MICCAI 2011 Workshop on Multi-Atlas Labeling and Statistical Fusion, Toronto, Canada, September 2011
Manifold Classification
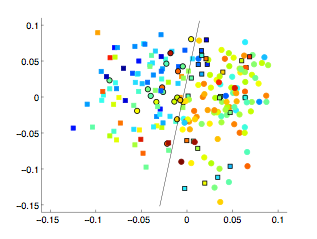
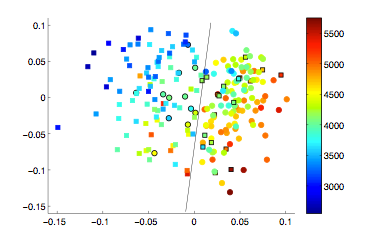
Learning a low-dimensional manifold space of a set of images can help to characterise the population of interest. If the clinical status of some subjects is available, it can be used to infer on the status of unlabelled subjects. In this work, the low-dimensional manifold space learned from subjects with Alzheimer’s disease and healthy controls is used to identify subjects at risk of developing the disease. Ways are proposed to incorporate subjects metadata and longitudinal MRI into the manifold learning process, increasing the accuracy of the learned space and leading to a better prediction accuracy.
Related Publications:
-
• R. Wolz, P. Aljabar, J.V. Hajnal and D. Rueckert
Manifold learning for biomarker discovery in MR imaging
MICCAI 2010 Workshop on Machine Learning in Medical Imaging, Beijing, China, September 2010
-
• R. Wolz, P. Aljabar, J. V. Hajnal, J. Lotjonen, D. Rueckert
Manifold Learning Combining Imaging with Non-Imaging Information
ISBI 2011, Chicago, USA, April 2011 (Finalist Best Student Paper Award)
-
• R. Wolz, P. Aljabar, J. V. Hajnal, J. Lotjonen, D. Rueckert
Manifold-based classification incorporating subject metadata
MIUA 2011, London, UK, July 2011