
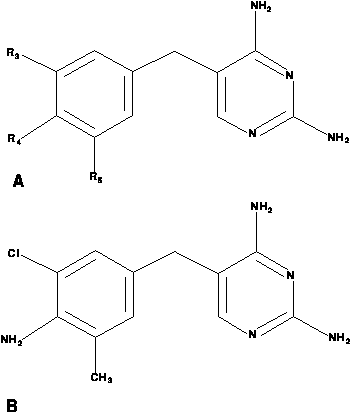
The existence of a template with only three possible substitution positions gives the pyrimidine problem a relatively small structural component. The chemical structure of the example compound above is adequately represented by a Prolog fact of the form
struc( d55, cl, nh2, ch3 )which is intended to represent that drug 55 has a chlorine atom substituted at position 3, an amine group (NH2) group substituted at position 4, and a methyl (CH3) group at position 5.
In [King R.D, Muggleton S., and Sternberg M.J.E. (1992)], the authors use 9 integer-valued attributes to represent chemical properties of the substituents. These were encoded as predicates of the form:
polar( br, polar3 )which states that a bromine atom has a polarity of value 3.
Positive examples are pairs of drugs, the activity of one being known to be higher than the other. For example:
great( d1, d2 )states that the E. Coli Dihydrofolate Reductase inhibition by drug
d1
is higher than that by d2.
In the following diagram of triazine structures,
the first structure is a generic template
for all compounds in the study, and the second is an example with
3=Cl, 4=(CH2)2 C6H3-4-Cl, 5=CH3
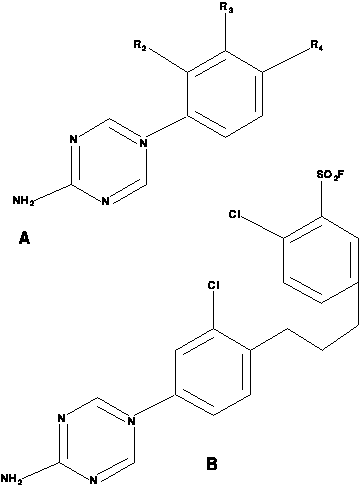
The first-order representation of the triazines is best explained using an example. The example compound above is represented by the following Prolog facts:
struc3( d217,cl, absent ). struc4( d217, '(ch2)4', subst14 ). subst( subst14, so2f, cl ).
The first clause represents substitutions at position 3 on the basic template: a
Cl is present and there is an absence of a further phenyl ring. The second
clause represents substitutions at position 4 on the basic template: there is a
(CH2)4 bridge to a second phenyl ring (implicit in the representation). This
second phenyl ring has an S02F group substituted at position 3 and a Cl group
substituted at position 4. This is represented using the linker constant
subst14
to the third clause. There is no substitution at position 2 on the basic
template. Each of the chemical groups had 10 attributes, 9 of which were the same
as used in the study of pyrimidines. One further attribute
was added to capture flexibility of a substituent. The degree of
flexibility is represented by one of 9 values.
King R.D, Muggleton S., and Sternberg M.J.E. (1992).
Drug design by machine learning: The use of inductive logic programming to model the structure-activity relationships of trimethoprim analogues binding to dihydrofolate reductase.
Proc. of the National Academy of Sciences, 89(23):11322--11326,
King, R.D., Srinivasan, A. and Sternberg, M.J.E. (1995).
Relating chemical activity to structure: an examination of ILP successes.
New Gen. Comput. (to appear).
 Machine Learning Group Home Page
Machine Learning Group Home Page