
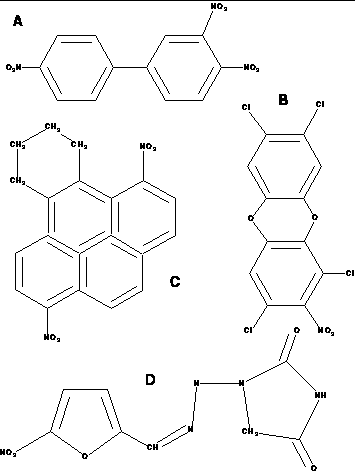
This figure shows examples of compounds
used in the mutagenesis study.
(A) 3,4,4'-tri-nitro-biphenyl
(B) 2-nitro-1,3,7,8-tetrachlorodibenzo-1,4-dioxin
(C) 1,6,-dinitro-9,10,11,12-tetrahydrobenzo[e]pyrene
(D) nitrofurantoin
The data here comes from ILP experiments conducted with Progol. Results of interest to the Machine Learning community are available in [Srinivasan, A., Muggleton, S., King, R.D., and Sternberg, M.J.E. (1994)], [Srinivasan, A., Muggleton, S.H., Sternberg, M.J.E., and King, R.D. (1995)], and [Srinivasan, A., Muggleton, S.H., Sternberg, M.J.E., and King, R.D. (1995)]. Relevant chemical results are in [King, R.D., Muggleton, S.H., Srinivasan, A., and Sternberg, M.J.E. (1995)]. Of the 230 compounds, 138 have positive levels of log mutagenicity, these are labelled "active" and constitute the positive examples: the remaining 92 compounds are labelled "inactive" and constitute the negative examples. Of course, algorithms that are capable of full regression can attempt to predict the log mutagenicity values directly.
The original Debnath et al paper recognised two subsets of data: 188 compounds that could be fitted using linear regression, and 42 compounds that could not. For the Progol experiments, accuracies of theories constructed for the 188 compounds were estimated from a 10-fold cross-validation. The accuracy of theories for the 42 compounds were estimated by a leave-one-out procedure.
The ILP experiments used the obvious generic description of compounds consisting of atoms and their bond connectivities. The compounds were input into the molecular modelling program QUANTA using its chemical editing facility. QUANTA then automatically adds typing information and calculates approximate partial charges associated with each atom. The choice of QUANTA was arbitrary, any similar molecular modelling package would have been suitable. The result is that each compound is represented by a sets of facts of the form:
atm( 127, 127_1, c, 22, 0.191 ) bond(127, 127_1, 127_6, 7 )
These two predicates give, in conjunction with ILP, the first completely generic method of describing molecular structure in drug design. The predicates also allow a straightforward definition of generic chemistry knowledge. This knowledge takes the form of a series of Prolog programs that define higher level chemical concepts (for example, ring structures).
In , four attributes are provided for analysis of the compounds. These can be used directly by both propositional and ILP learners. They are:
Debnath, A.K. Lopez de Compadre, R.L., Debnath, G., Shusterman, A.J., and Hansch, C. (1991).
Structure-activity relationship of mutagenic aromatic and heteroaromatic nitro compounds. Correlation with molecular orbital energies and hydrophobicity.
J. Med. Chem. 34:786-797.
King, R.D., Muggleton, S.H., Srinivasan, A., and Sternberg, M.J.E. (1995)
Representing molecular structure in structure activity relationships: The use of atoms and their bond connectivities to predict mutagenicity using inductive logic programming.
Submitted to J. Am. Chem. Soc.
Srinivasan, A., Muggleton, S., King, R.D., and Sternberg, M.J.E. (1994)
Mutagenesis: ILP experiments in a non-determinate biological domain.
Proceedings of the Fourth Inductive Logic Programming Workshop.
Srinivasan, A., Muggleton, S.H., Sternberg, M.J.E., and King, R.D. (1995)
Theories for mutagenicity: a study of first-order and feature based induction.
PRG-TR-8-95, Oxford University Computing Laboratory.
Srinivasan, A., Muggleton, S.H., Sternberg, M.J.E., and King, R.D. (1995)
The effect of background knowledge in Inductive Logic Programming:
a case study.
PRG-TR-9-95, Oxford University Computing Laboratory.
 Machine Learning Group Home Page
Machine Learning Group Home Page