
Predicting protein secondary structure
Contents and links
Predicting the three-dimensional shape of proteins
from their amino acid sequence is widely believed
to be one of the hardest unsolved problems in
molecular biology. It is also of considerable
interest to pharmaceutical companies since a
protein's shape
generally determines its function as an enzyme.
This is what a protein looks like.
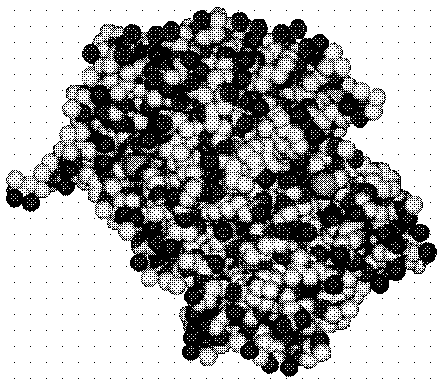 [Muggleton S., King R.D., and Sternberg M.J.E. (1992)].
The task is to learn rules to identify whether a position in a protein
is in an alpha-helix. Points of relevance are:
[Muggleton S., King R.D., and Sternberg M.J.E. (1992)].
The task is to learn rules to identify whether a position in a protein
is in an alpha-helix. Points of relevance are:
- Each amino acid is denoted by a lower case character. There are
20 such amino acids.
- Positive examples state which positions of chosen
proteins are in an alpha-helix. Negative examples
state the positions that are not in an alpha-helix.
- The following background knowledge is provided:
-
position(A,B,C). Residue of protein A
at position B is C.
-
octf(A,B,C,D,E,F,G,H,I). Arithmetic information that allows indexing
groups of nine adjacent positions in a protein.
Basically says positions A--I occur in sequence.
-
alpha_triplet(A,B,C). Arithmetic information that allows indexing
groups of three adjacent positions in a protein.
-
alpha_pair(A,B). Arithmetic information that allows indexing
a pair of adjacent positions in a protein.
-
alpha_pair4(A,B). Arithmetic information that allows indexing
a pair of positions separated by 4 positions in a protein.
- Physical and chemical properties of individual residues are described
by unary predicates. These properties include hydrophobicity, hydrophilicity,
charge, size, polarity, whether a residue is aliphatic or aromatic, whether
it is a hydrogen donor or acceptor etc.
- Sizes, hydrophobicities, polarities etc., are represented by constants
such as
polar0 and polar1.
- Relations between the constants (
less_than(polar0,polar1)) is
also provided as background knowledge.
The data files we provide are as used in the original Golem experiments,
and are downloadable as
one compressed TAR file.
Within this file, background knowledge files have a ``.b'' suffix,
positive example files have a ``.f'' suffix, and negative example files
have a ``.n'' suffix.
Bibliography
Muggleton S., King R.D., and Sternberg M.J.E. (1992).
Predicting protein secondary structure using inductive logic programming.
in Protein Engineering, 5:647--657.
Up to applications main page.
 Machine Learning Group Home Page
Machine Learning Group Home Page


 [Muggleton S., King R.D., and Sternberg M.J.E. (1992)].
The task is to learn rules to identify whether a position in a protein
is in an alpha-helix. Points of relevance are:
[Muggleton S., King R.D., and Sternberg M.J.E. (1992)].
The task is to learn rules to identify whether a position in a protein
is in an alpha-helix. Points of relevance are:
 Machine Learning Group Home Page
Machine Learning Group Home Page