Research
Graph Convolutional Neural Networks
Convolutional Neural Networks (CNNs) have found numerous applications on 2D and 3D images, as powerful models that exploit features (e.g. image intensities) and neighbourhood information (e.g. the regular pixel grid) to yield hierarchies of features and solve problems like image segmentation and classification. Their impressive performance can be partly linked to the fact that CNNs intrinsically exploit the relationship between neighbouring pixels through the images’ regular grid structure.
Applying similarly powerful tools for data defined on irregular graphical domains is not straightforward. In this project, we explore the novel concept of graph CNNs for brain analysis using population and brain graphs. In particular, spectral graph CNNs expoit the concept of signal processing on graphs, allowing convolutions in the graph spatial domain to be dealt as multiplications in the graph spectral domain.
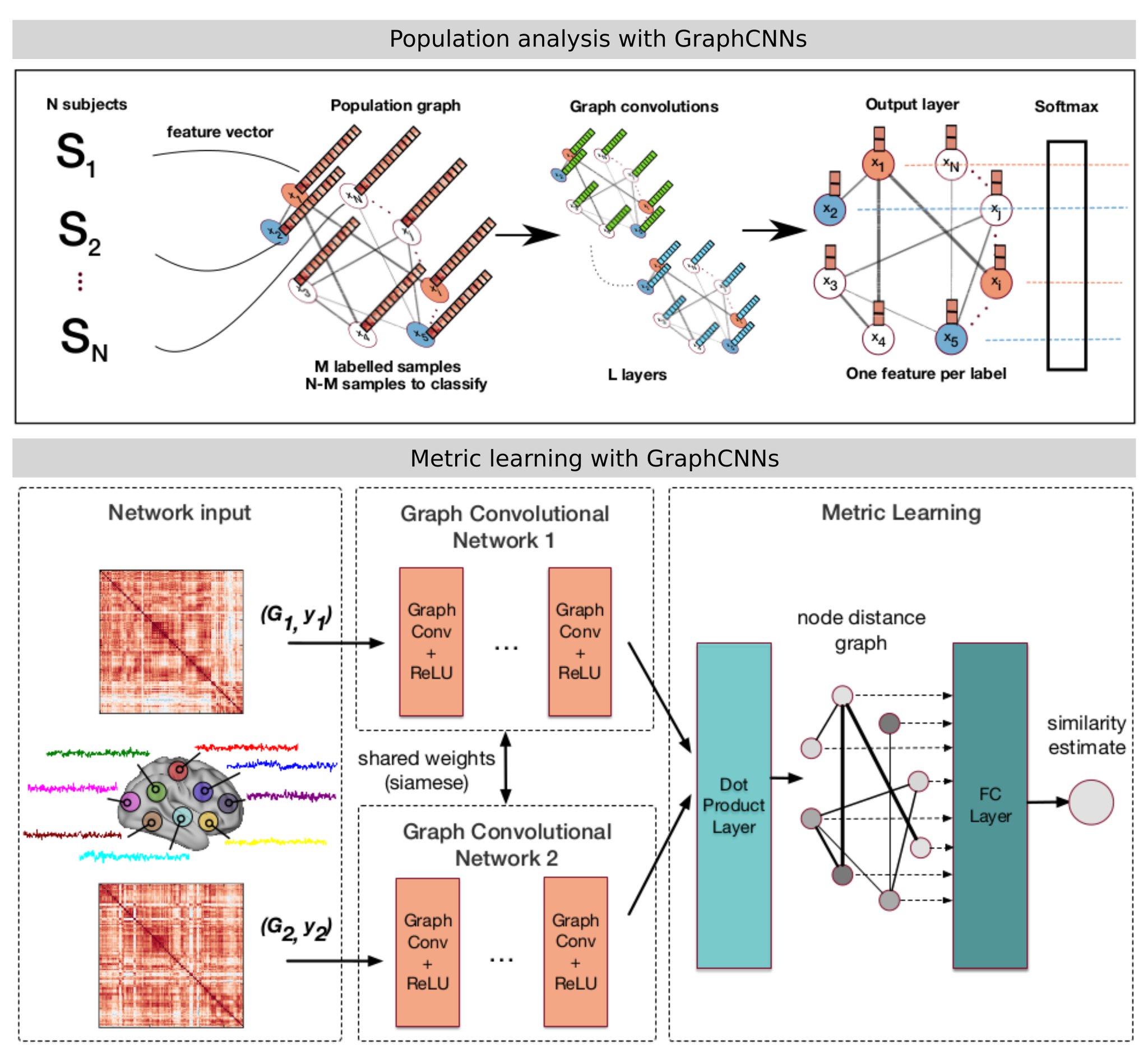
Such formulation was exploited for:
- Brain analysis in populations, combining imaging and non-imaging data through the construction of graphs representing populations. Semi-supervised classification of the graph nodes is carried out for diagnosis tasks.
- The comparison of brain graphs, using a siamese graph CNN to learn a similarity metric between brain connectivity networks.
Key methodological aspects:
- Machine/deep learning and applications to unstructured domains
- Graph signal processing
- Semi-supervised classification
- Metric learning
Selected publications: Parisot et al. MICCAI 2017 Ktena et al. MICCAI 2017
Connectivity-driven Brain Parcellation
This project is carried out under the scope of the Developing Human Connectome Project.

Brain connectivity (structural from diffusion MRI or functional from functional MRI) can provide essential information on the interactions between brain regions and their processing abilities. It notably has the potential to unravel the mechanisms behind neurodegenerative diseases or ageing.
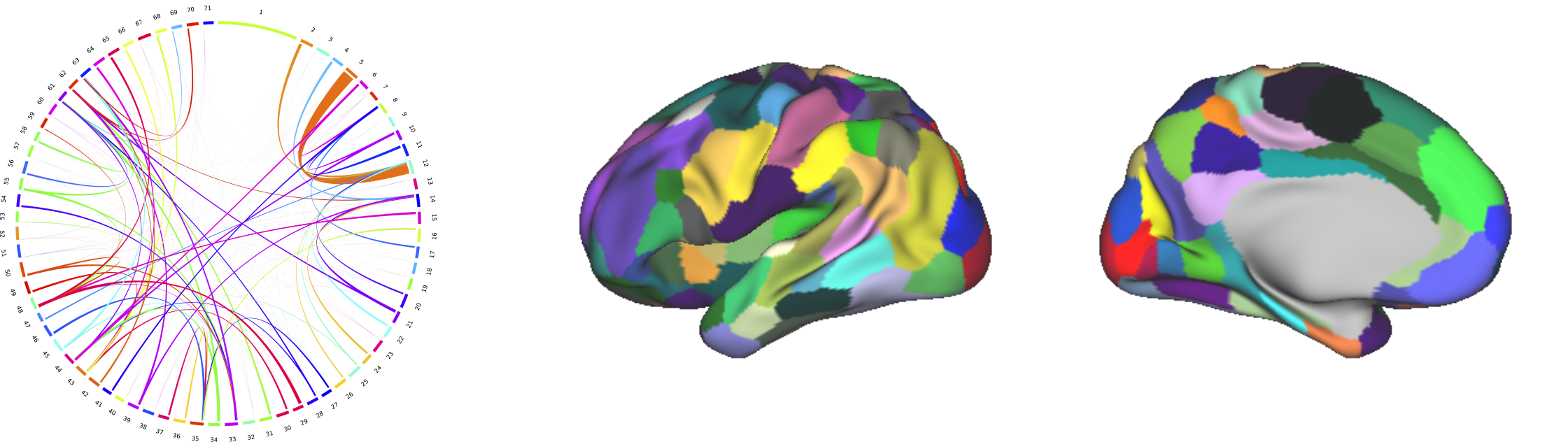
The goal of connectivity-driven brain parcellation is two-fold:
Dimensionality reduction: In vivo connectivity data is obtained at the voxel or vertex level. The dimensionality of this data is too high to extract any meaningful information from connectivity networks. We aim at identifying the nodes of the connectivity network at a lower resolution, where each node correspond to a brain region that has consistent connectivity profiles.
Identification of functionally specified regions: On top of building connectivity networks, regions that have sharp transitions of connectivity profiles can be an indicator of a different functional specialisation.
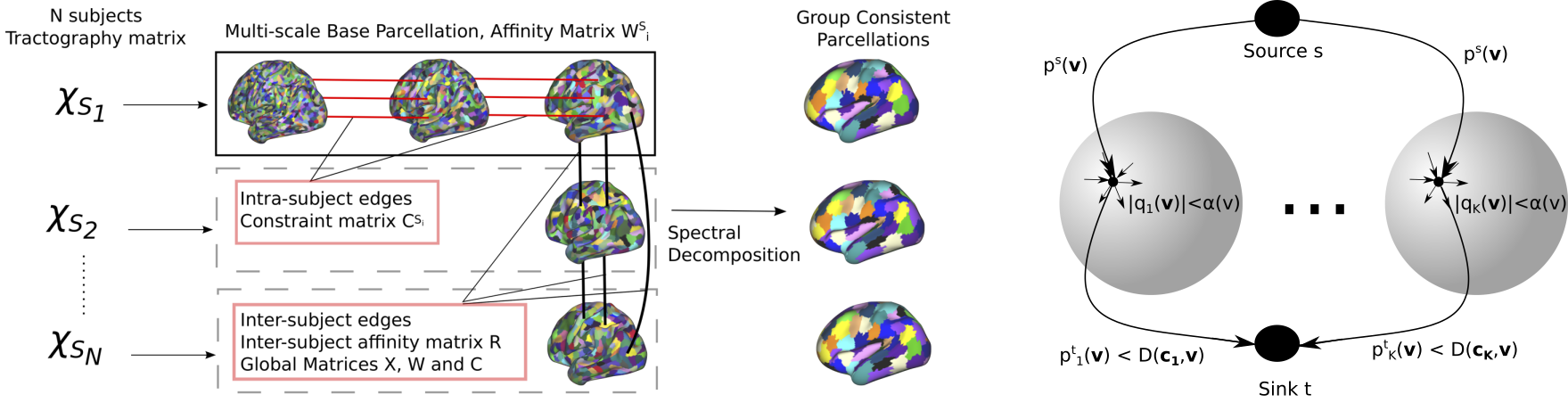
Several approaches were considered to tackle the task of brain parcellation:
- We investigated group-wise parcellation through spectral clustering techniques.
- Modelling the parcellation task as an iterative continuous Potts model using the recent continuous max-flow optimiser.
- The Potts model formulation was later extended for multi-modal parcellation and discrete optimisers.
Key methodological aspects:
- Multi-scale spectral clustering
- Continuous and Discrete Potts models
- Adaptation of a state of the art optimiser to spherical meshes
- Computer vision methods (co segmentation) for groupwise parcellation
Selected publications: Parisot et al. MICCAI 2016. [DOI][Bibtex][PDF] Parisot et al. NeuroImage (2016). [DOI] [Bibtex][PDF] Arslan et al. NeuroImage (2017)[DOI] [PDF]
Concurrent Registration/Segmentation
The idea behind this project was to exploit the codependency between segmentation and registration with missing correspondences. In typical sequential approaches, the missing object is first segmented and then masked in the registration process.
We proposed a flexible concurrent registration/segmentation method that makes no assumption of the registration similarity criterion or the object to be segmented. We applied the method on brain tumours and intra-operative registration with tumour resection.
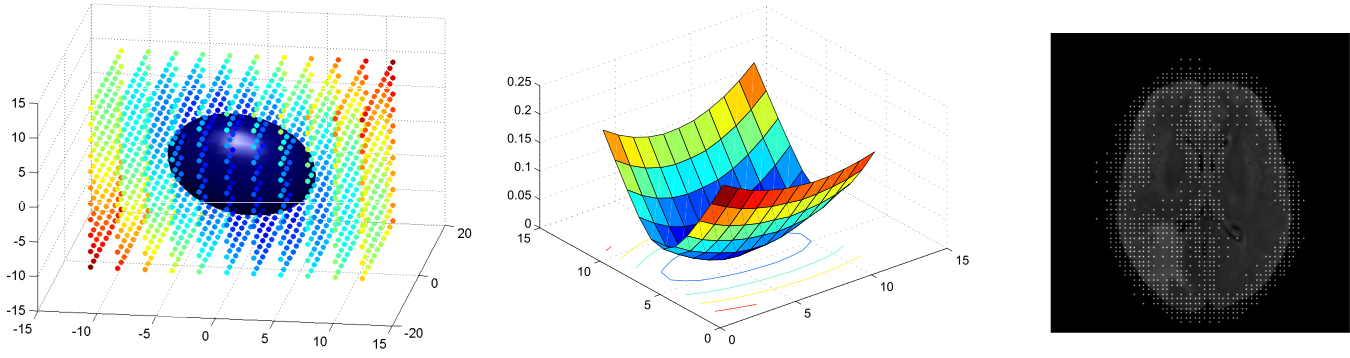
The method was extended for content-driven sparse sampling driven from registration and segmentation uncertainty. We aim at increasing the efficiency of our algorithm by building sparse non-uniform grids where the segmentation and registration parameters are evaluated.
Key methodological aspects:
- Registration with missing correspondences
- Atlas-based segmentation
- Content-driven sparse sampling
- Registration and segmentation uncertainty
Related publications: Parisot et al. MICCAI 2012 [DOI] [Bibtex] [PDF] [Slides] Parisot et al. ICCV 2013 [DOI] [Bibtex] [PDF] Parisot et al. Medical Image Analysis 2014 [DOI] [Bibtex] [PDF]
Atlas-based Brain Tumour Segmentation
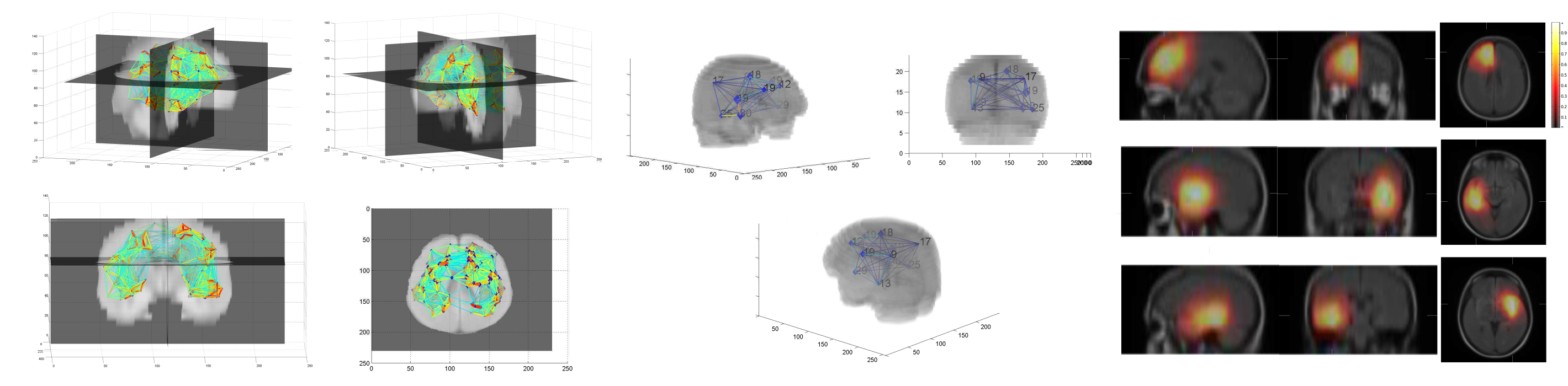
Brain tumour segmentation was the main objective of my PhD thesis. Instead of focusing on machine learning methods which remain dependent on the appearance of the image pixels, we focused on incorporating high level information in the segmentation task.
We built a probabilistic atlas describing the tumour’s preferential locations, which was integrated in a graph-cut segmentation algorithm as a position prior.
Key methodological aspects:
- Network analysis and clustering
- Markov Random Field models
- Atlas-based segmentation
Related publications: Parisot et al. MICCAI 2011 [DOI] [Bibtex] [PDF] Parisot et al. CVPR 2012 [DOI] [Bibtex] [PDF]

