Dr Alireza Tamaddoni Nezhad |
 |
|
Address: Google Scholar Profile Research Gate Profile |
 |
Dr Alireza Tamaddoni Nezhad |
 |
|
Address: Google Scholar Profile Research Gate Profile |
 |
1. Automated
discovery of food-webs using machine learning and
text-mining
| I have developed [30, 10] a machine learning method which can be used to generate probabilistic interaction networks from ecological data. The data used in this project was sampled from 257 fields across the UK in the Farm Scale Evaluations (FSE) of GM, herbicide tolerant (GMHT) crops. The initial food-webs proposed by machine learning were examined [13] by experts from Rothamsted Research UK and it was found that many of the learned trophic links were corroborated by the literature. In particular, links ascribed with high probability by machine learning were shown to correspond well with those having multiple references in the literature. In some cases novel, high probability links were suggested, and some of these have been tested and confirmed by subsequent empirical studies (e.g. Davey et. al., 2013) and DNA analysis. This study is regarded as the first successful application of machine learning from large scale agricultural data and the machine learning approach and the analysis of the learned trophic links were published in high impact journals (e.g. [10]). | 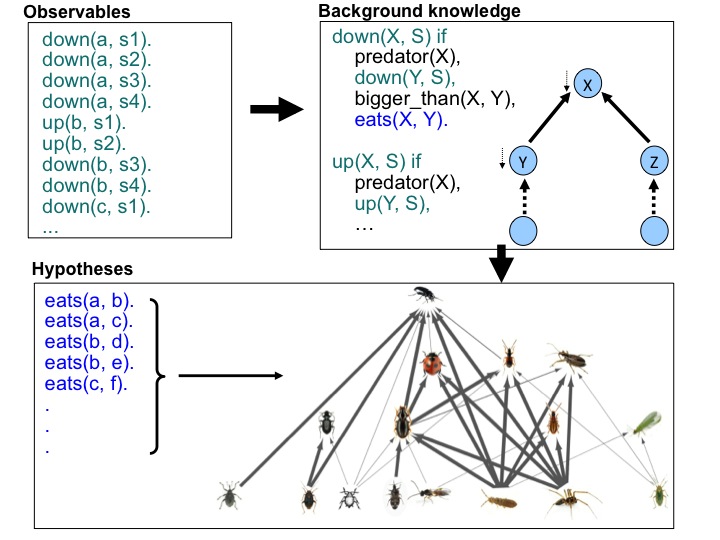 |
2.
Integrative systems biology of bacterial glycomes: modelling
genotype-phenotype relations in Campylobacter
| In this project a mixture of metabolic data, gene expression and mutants data were used together to model genotype-phenotype relations in biosynthetic pathways for capsular polysaccharides of Campylobacter jejuni. The experimental observations were the capsule phenotypes of gene knockout mutants using HR-MAS NMR data. The aim of the project was to discover new gene functions in order to develop novel treatment against diseases caused by Campylobacter. In this project a logic-based machine learning approach was developed [12] to provide a robust strategy for integrating data from different experimental sources. | 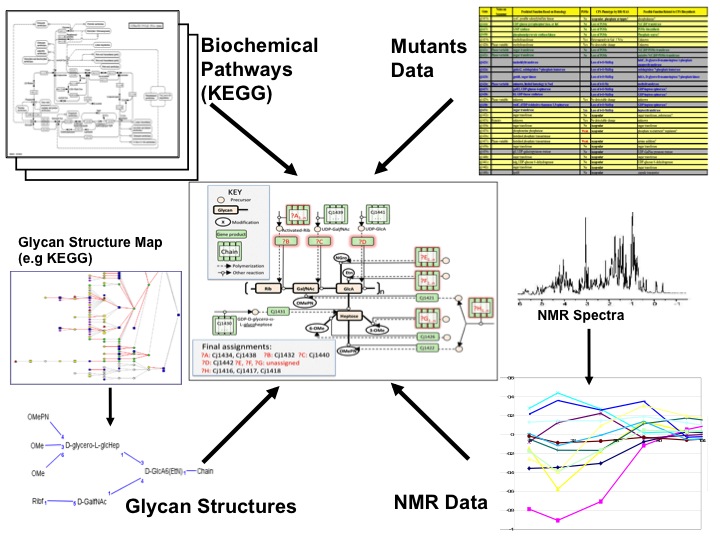 |
3. MetaLog - integrated
machine learning of metabolic networks applied to predictive
toxicology
| The main aim of this project was to provide causal explanations of rat liver cell responses to toxins (e.g. Hydrazine). The observational data consisted of up and down regulation patterns found in high-throughput NMR data collected by the Consortium for Metabonomic Toxicology (COMET) formed between six pharmaceutical companies and Imperial College. This study [18,16] is regarded as a successful application of relational and logic-based machine learning for modelling metabolic networks and the method developed in this project was used in several follow-up studies by other researchers. | 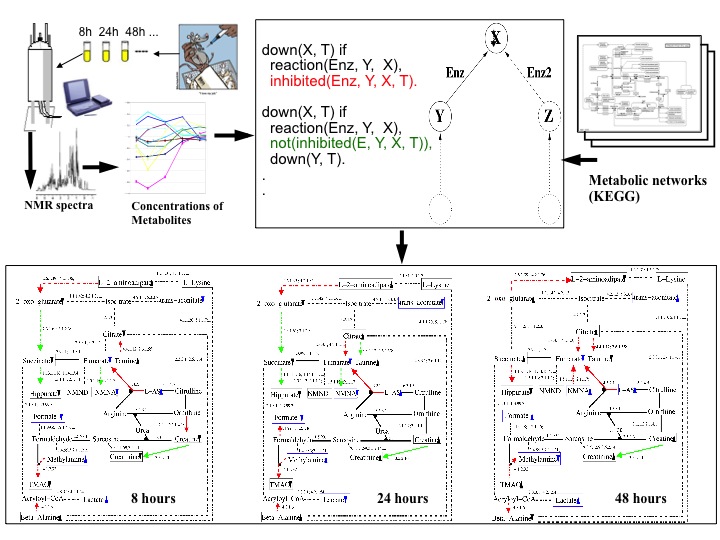 |
4.
Logic-base
machine learning using a bounded hypothesis space: lattice
structure, refinement operators and a genetic algorithm
approach
| During my PhD study under the supervision of Prof. Stephen Muggleton, I analysed the lattice structure and refinement operators for the bounded hypothesis space in Inductive Logic Programming (ILP) and introduced the concept of bounded subsumption (i.e. subsumption relative to a bottom clause) [15]. I also developed novel stochastic search methods including a Genetic Algorithm approach for ILP [17, 32, 45, 46, 47]. The bounded hypothesis is the basis of several state-of-the art ILP systems (e.g. ProGolem, Progol and Aleph) which have been applied to many real-world problems. In my PhD I have also shown that the bounded subsumption lattice is isomorphic to atomic lattice and that the general subsumption testing, which is NP-complete, can be reduced to unification which has a linear time complexity. The Stochastic Refinement approach [32] introduced in my PhD, has been adapted [27] for Bayesian Meta-Interpretive Learning (MetaBayes) which is shown to be useful in challenging problems in machine learning (e.g. learning from large scale textual data). | 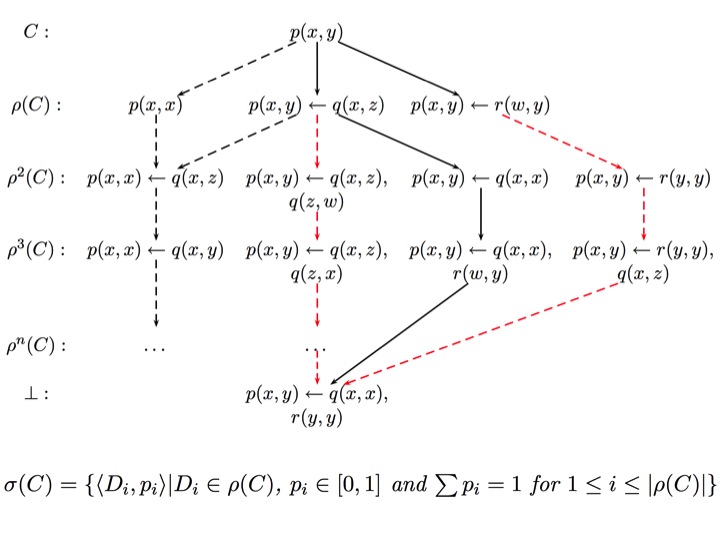 |
Journal Articles
[1] D. A. Ma, X. Lu, C. Gray, A. Raybould, A.
Tamaddoni-Nezhad, G. Woodward, D. Bohan. Ecological
networks reveal resilience of agro-ecosystems to changes
in farming management. Nature Ecology &
Evolution, 3:260-264, 2019
[2] S.H.
Muggleton, W-Z. Dai, C. Sammut, A. Tamaddoni-Nezhad, J.
Wen and Z-H. Zhou. Meta-interpretive
learning from noisy images. Machine Learning,
107:1097-1118, 2018.
[3] S.H.
Muggleton, U. Schmid, C. Zeller, A. Tamaddoni-Nezhad, and
T. Besold. Ultra-strong
machine learning - comprehensibility of programs learned
with ILP. Machine Learning, 107:1119-1140,
2018
[4] D. Bohan, C. Vacher, A. Tamaddoni-Nezhad, A. Raybould, A. Dumbrell and G. Woodward, Next- Generation Global Biomonitoring – Large-scale, automated reconstruction of ecological networks. Trends in Ecology and Evolution, 32(7), 477-487, 2017
[5] C. Vacher, A. Tamaddoni-Nezhad,
S. Kamenova, N. Peyrard,
Y. Moalic, R. Sabbadin, L. Schwaller, J. Chiquet, M. Smith, J. Vallance, V. Fievet, B. Jakuschkin, D. A. Bohan, Learning
Ecological Networks from Next-Generation Sequencing Data.
Advances in Ecological Research, 54, 1-39, 2016.
[6] M. Pocock, D. Evans, C. Fontaine, M.
Harvey, R. Julliard, Ó. McLaughlin, J. Silvertown, A.
Tamaddoni-Nezhad, P. White, and D. Bohan, The visualisation
of ecological networks, and their use as a tool for
engagement, advocacy and management, Advances in
Ecological Research, 54, 1-39, 2016.
[7] QUINTESSENCE Consortium. Networking our way to
better Ecosystem Service provision. Trends in Ecology and
Evolution, 31(2):105-115, 2016.
[8] S.H. Muggleton, D. Lin and
A. Tamaddoni-Nezhad. Meta-interpretive
learning of higher-order dyadic datalog: predicate invention
revisited, Machine
Learning, 100(1):49-73,
2015.
[9] S.H. Muggleton, D. Lin, N. Pahlavi, and A. Tamaddoni-Nezhad. Meta-Interpretive Learning: application to Grammatical Inference. Machine Learning, 94:25-49, 2014
[10] A. Tamaddoni-Nezhad, G. Afroozi Milani, A. Raybould, S. Muggleton and D.Bohan, Construction and Validation of Agricultural Food-webs using Logic-based Machine Learning and Text-mining, Advances in Ecological Research, Vol 49, pages 224-290, Academic Press, Amsterdam, 2013
[11] D. Bohan, A. Raybould, C. Mulder, G. Woodward, A. Tamaddoni-Nezhad, N. Bluthgen, M. Pocock, S. Muggleton, D. Evans, J. Astegiano, F. Massol, N. Loeuille, S. Petit and S. Macfadyen. Networking agroecology: Integrating the diversity of agroecosystem interactions Advances in Ecological Research, Vol 49, pages 2-67. Academic Press, Amsterdam, 2013
[12] M. Sternberg, A.
Tamaddoni-Nezhad, V. Lesk, E. Kay, P. Hitchen, A. Cootes, L.
Alphen, M. Lamoureux, H. Jarrell, C. Rawlings, E. Soo, C.
Szymanski, A. Dell, B. Wren, S. Muggleton. Gene
function
hypotheses for the Campylobacter jejuni glycome generated by
a logic-based approach. Journal
of Molecular Biology,
425(1):186-197, 2013
[13] D. A. Bohan, G. Caron-Lormier, S. Muggleton, A. Raybould and A. Tamaddoni-Nezhad. Automated Discovery of Food Webs from Ecological Data Using Logic-Based Machine Learning. PloS One, vol. 6, pp. e29028, 2011.
[14] E Kay, V Lesk, A. Tamaddoni-Nezhad, P Hitchen, A Dell, M. Sternberg, S. Muggleton, B Wren. Systems analysis of bacterial glycomes. Biochemical Society Transactions, volume 38, issue 5, pp.1290–1293, 2010.
[15] A. Tamaddoni-Nezhad and S.H. Muggleton. The lattice structure and refinement operators for the hypothesis space bounded by a bottom clause. Machine Learning, 76(1):37-72, 2009.
[16] A. Tamaddoni-Nezhad, R. Chaleil, A. Kakas, M. Sternberg, J. Nicholson, and S. Muggleton. Modeling the effects of toxins in metabolic networks. IEEE Engineering in Medicine and Biology, 26:37-46, 2007.
[17] S.H. Muggleton and A.Tamaddoni-Nezhad. QG/GA: A stochastic search approach for Progol. Machine Learning, 70 (2–3):123–133, 2007. (Best Paper Award)
[18] A. Tamaddoni-Nezhad, R.
Chaleil, A. Kakas, and S.H. Muggleton. Application
of
abductive ILP to learning metabolic network inhibition from
temporal data. Machine
Learning, 64:209–230,
2006.
Book Chapter
[19] A. Tamaddoni-Nezhad, D. Lin, H. Watanabe, J.
Chen and S. Muggleton, Machine
Learning
of
Biological
Networks using Abductive ILP, In Eds. L. Cerro and K.
Inoue, Logical Modeling of Biological Systems, pp 363-401,
ISTE-Wesley, 2014
Edited Proceedings
[20] S.H. Muggleton , A. Tamaddoni-Nezhad & F.A. Lisi (Eds.) Inductive Logic Programming, 21st International Conference, ILP 2011, Revised Selected Papers, Series: Lecture Notes in Computer Science, Vol. 7207, Springer, 2012
[21] S.H. Muggleton, R. Otero, and A. Tamadonni-Nezhad (Eds.) Inductive Logic Programming, 16th International Conference, ILP 2006, Revised Selected Papers, Series: Lecture Notes in Computer Science, Vol. 4455, Springer, 2007.
Refereed Conference and Workshop Papers
[22] W-Z
Dai, S.H. Muggleton, J. Wen, A. Tamaddoni-Nezhad, and Z-H.
Zhou. Logical vision: One-shot meta-interpretive learning
from real images. In Proceedings of the 27th International
Conference on Inductive Logic Programming,
Springer-Verlag, pages 46-62, 2018.
[23] U. Schmid, C. Zeller, T. Besold, A. Tamaddoni-Nezhad, and S.H. Muggleton. How does predicate invention affect human comprehensibility? In Proceedings of the 26th International Conference on Inductive Logic Programming, Springer-Verlag, pages 52-67, 2017.
[24] W. Dai , S. Muggleton , A.
Tamaddoni-Nezhad and Z. Zhou, Logical
vision: meta-interpretive learning for human-like vision, Human-Like
Computing Machine Intelligence Workshop (MI20- HLC), 2016
[25] A. Cropper, A. Tamaddoni-Nezhad, and S. Muggleton. Meta-interpretive learning of data transformation programs. In Proceedings of the 25th International Conference on Inductive Logic Programming. Springer-Verlag, pages 46-59, 2016.
[26] A.
Tamaddoni-Nezhad, D. Bohan, A. Raybould and S. Muggleton.
Towards machine learning of predictive models from ecological
data. In Proceedings of the 24th International Conference
on Inductive Logic Programming, Springer-Verlag, pages
154-167, 2015.
[27] S.H. Muggleton, D. Lin,
J. Chen, and A. Tamaddoni-Nezhad. Metabayes:
Bayesian meta-interpretative learning using higher-order
stochastic refinement. In Proceedings of the 23rd International Conference
on Inductive Logic Programming, LNAI 8812, pages 1-17, 2014
[28] A. Tamaddoni-Nezhad, G. Afroozi Milani, D. Bohan, S. Dunbar, A. Raybould, and S.H. Muggleton. Towards automatic construction and corroboration of food webs. In Proceedings of the European Conference on Machine Learning, Workshop on Learning and Discovery in Symbolic Systems Biology (ECML/LDSSB 2012), pages 95-102, 2012
[29] G. Afroozi Milani, D. Bohan, S. Dunbar, A. Raybould, S.H. Muggleton and A. Tamaddoni-Nezhad, Machine learning and text mining of trophic links. In Proceedings of the 11th International Conference on Machine Learning and Applications Special Session on Learning on the Web (ICMLA/LW 2012), pages 410-415, IEEE, 2012
[30] A. Tamaddoni-Nezhad, D. Bohan, A. Raybould and S. Muggleton. Machine learning a probabilistic network of ecological interactions. In Proceedings of the 21st International Conference on Inductive Logic Programming, LNAI 7207, pages 332-346, 2012. (Best Paper Award)
[31] S.H. Muggleton, D. Lin, and A. Tamaddoni-Nezhad. MC-Toplog: Complete multi-clause learning guided by a top theory. In Proceedings of the 21st International Conference on Inductive Logic Programming, LNAI 7207, pages 238-254, 2012.
[32] A. Tamaddoni-Nezhad and S.H. Muggleton. Stochastic Refinement. In Proceedings of the 20th International Conference on Inductive Logic Programming, pages 222-237, 2011.
[33] S.H. Muggleton, J. Santos, and A. Tamaddoni-Nezhad. ProGolem: a system based on relative minimal generalisation. In Proceedings of the 19th International Conference on Inductive Logic Programming, LNCS 5989, pages 131-148. Springer-Verlag, 2010.
[34] S.H. Muggleton, J. Santos, and A. Tamaddoni-Nezhad. TopLog: ILP using a logic program declarative bias. In Proceedings of the International Conference on Logic Programming, LNCS 5366, pages 687-692. Springer-Verlag, 2010.
[35] A. Tamaddoni-Nezhad, R. Barton, P. Hitchen, E. Kay, V. Lesk, F. Turner, A. Dell, C. Rawlings, M. Sternberg, B. Wren, S. Muggleton, A logic-based approach for modeling genotype-phenotype relations in Campylobacter, In Proceedings of the 9th International Conference on Systems Biology (ICSB-2008), 2008.
[36] J. Santos, A. Tamaddoni-Nezhad, and S.H. Muggleton. An ILP system for learning head output connected predicates. In Proceedings of the 14th Portuguese Conference on Artificial Intelligence, LNAI 5816, pages 150-159. Springer-Verlag, 2009.
[37] A. Tamaddoni-Nezhad and S.H. Muggleton. A note on refinement operators for IE-based ILP systems. In Proceedings of the 18th International Conference on Inductive Logic Programming, LNAI 5194, pages 297-314. Springer-Verlag, 2008.
[38] S.H. Muggleton and A. Tamaddoni-Nezhad. QG/GA: A stochastic search approach for Progol. In Proceedings of the 16th International Conference on Inductive Logic Programming, LNAI 4455, pages 37-39. Springer-Verlag, 2006.
[39] A. Tamaddoni-Nezhad, R. Greaves, and S.H. Muggleton. Large-scale online learning using analogical prediction. In Short Paper Proceedings of the 16th International Conference on Inductive Logic Programming. University of Corunna, 2006.
[40] A. Tamaddoni-Nezhad, R. Chaleil, A. Kakas, S.H. Muggleton. Abduction and induction for learning models of inhibition in metabolic networks. In Proceedings of the International Conference on Machine Learning and Applications (ICMLA’05), pages 233-238, IEEE. 2005.
[41] G. Afroozi Milani, K. Ziarati, and A. Tamaddoni-Nezhad. Virtual characteristics measurement using factor analysis. In Proceedings of the 2nd International Conference on E-Business and Telecommunication Networks (ICETE'05), pages 102-108, 2005.
[42] A. Tamaddoni-Nezhad, A. Kakas, S.H. Muggleton, and F. Pazos. Modelling inhibition in metabolic pathways through abduction and induction . In Proceedings of the 14th International Conference on Inductive Logic Programming, pages 305-322. Springer-Verlag, 2004.
[43] S.H. Muggleton, A. Tamaddoni-Nezhad, and H. Watanabe. Induction of enzyme classes from biological databases. In Proceedings of the 13th International Conference on Inductive Logic Programming, pages 269-280. Springer-Verlag, 2003.
[44] A. Tamaddoni-Nezhad, S. Muggleton, and J. Bang. A Bayesian model for metabolic pathways. In International Joint Conference on Artificial Intelligence (IJCAI03) Workshop on Learning Statistical Models from Relational Data, pages 50-57. IJCAI, 2003.
[45] A. Tamaddoni-Nezhad and S.H. Muggleton. A genetic algorithms approach to ILP. In Proceedings of the 12th International Conference on Inductive Logic Programming, pages 285-300. Springer-Verlag, 2002.
[46] A. Tamaddoni-Nezhad and S.H. Muggleton. Using genetic algorithms for learning clauses in first-order logic. In Proceedings of the Genetic and Evolutionary Computation Conference, GECCO-2001, pages 639-646, Morgan Kaufmann Publishers, 2001.
[47] A. Tamaddoni-Nezhad
and S.H. Muggleton. Searching
the subsumption lattice by a genetic algorithm. In Proceedings of the 10th International Conference on
Inductive Logic Programming, pages 243-252. Springer-Verlag, 2000.
Technical Reports
[48] A Tamaddoni-Nezhad and S Muggleton. Closed loop Machine Learning: Complexity of ASE-Progol. Robot Scientist Project, TR 2002/8, Dept. of Computing, Imperial College London, 2002.
[49] A Tamaddoni-Nezhad
and S Muggleton. Closed loop Machine Learning:
Reproduction and evaluation of phase A results. Robot
Scientist Project, TR 2002/7, Dept. of Computing, Imperial
College London, 2002.